APE SCHTICK: Scientists Create World’s Largest Chimp DNA Genome From Poop
An international team of experts has compiled the world’s largest wild chimp genomic catalogue by sequencing DNA found in hundreds of samples of ape poo.
The team was led by the Institute of Evolutionary Biology (IBE), the German Centre for Integrative Biodiversity Research (iDiv), the Max Planck Institute for Evolutionary Anthropology (MPI EVA) and Leipzig University.
A statement obtained from iDiv revealed that the experts had harvested genomic information from hundreds of chimp faecal samples.

They say the DNA map could clear up questions about apes’ evolutionary past and be used to crack down on poachers and traffickers.
The iDiv statement said: “The new genomic atlas, published in the journal Cell Genomics, has the potential to map routes and sources of illegal trafficking that could be used to protect this endangered species.”
They explained that Chimpanzees live in woodlands and forests in the tropical African savannah and that unlike ancient human sites, primarily in caves and temperate climates, the places where chimps live have meant that there are very few “specimens preserved or detected in the archaeological record”.

The experts said: “Given the almost complete absence of chimpanzee fossils, the genetic information from current populations is crucial for describing their evolutionary history and their genetic diversity and contributing to their conservation.”
The statement added: “Genetic information has been retrieved using new technologies, from hundreds of non-invasively collected chimpanzee faecal samples.
“For the first time, methods applied to analyse ancient DNA in human populations have been used to retrieve genetic information from great apes’ faecal samples.

“Further, the genomic database developed by the team can be used in practice for the conservation of chimpanzees, such as identifying illegal trafficking routes of wildlife products and orphans.”
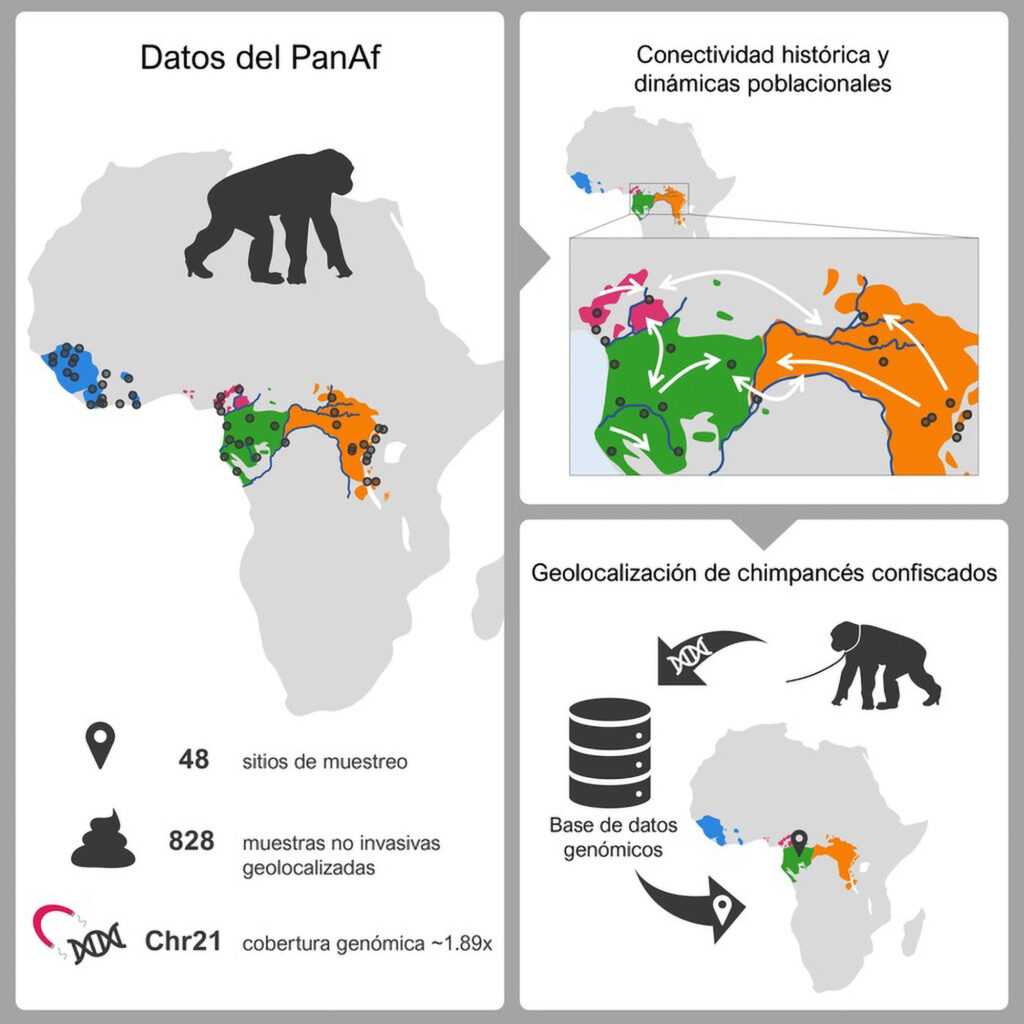
By harvesting more than 800 chimp poo samples, the research could help map out migratory patterns going back 100,000 years, say the experts.
Professor Tomas Marques-Bonet, of the IBE and a co-author of the study, said: “Using methods designed to study ancient DNA, as in the case of Neanderthals, we have been able to retrieve genomic information from faecal samples, which are very difficult to work with.
“We have applied that approach to an unprecedented number of chimpanzee samples from the field.”
Dr Claudia Fontsere, a researcher at the IBE Comparative Genomics group and the first author of the study, said: “We have seen that faecal samples, while posing technical difficulties, provide very valuable genomic information for the study of wild chimpanzee populations, and also allow us the possibility to geo-reference them and track contacts between populations without affecting their well-being.”
She added: “We’ve noticed that sometimes, even though two communities are geographically very close, they may live on two different sides of a river and have only had very limited and sporadic contact.

“Our approach is very helpful in identifying barriers and natural corridors between populations and may have implications for their conservation.”
Co-author Dr Mimi Arandjelovic, a researcher at iDiv, MPI EVA and Leipzig University, who has spent eight years collecting behavioural, ecological and organic data from across the entire chimpanzee range, said: “Chimpanzees, like humans, have a complex evolutionary history.
“Their dynamics and areas of past and current population contact must be clearly identified in order to contribute to the protection of this endangered species.”
Dr Arandjelovic is also the co-director of the Pan African Programme: The Cultured Chimpanzee (PanAf), a consortium of researchers and conservationists from Africa, Europe and North America
Chimpanzees are listed as endangered on the International Union for Conservation of Nature’s (IUCN) Red List of Threatened Species.
The new tool will help researchers work out where individuals came from originally, which was not possible until now.

The statement said: “The new genomic tool has allowed the team to reliably identify from where individuals originate; a task not feasible until now.
“The ability to accurately determine the origin of chimpanzees has direct applications for their conservation, such as detecting the places where their poaching might be concentrated and identifying the routes and origins of illegal chimpanzee trafficking.”
Marques-Bonet added: “The tool developed can infer the origin of the confiscated chimpanzees, which are usually found just a few hundred kilometres from their real origin, and thus provide reliable information on the regions most in need of protection.”



